Back in the day, and I mean back in the day, material design was a labor-intensive endeavor. Researchers have been trying to create gold for over 1,000 years by mixing lead, mercury, sulfur, and more in the right proportions. Even famous scientists like Tycho Brahe, Robert Boyle, and Isaac Newton tried their hand at a futile endeavor called alchemy.
Of course, materials science has come a long way. For the past 150 years, researchers have benefited from the periodic table of elements. The table teaches researchers that different elements have different properties and that one element does not magically transform into another. Moreover, over the past decade or so, machine learning tools have greatly improved our ability to determine the structure and physical properties of various molecules and materials. A new study by a team led by Ju Lee, MIT Professor of Nuclear Engineering and Professor of Materials Science and Engineering at Tokyo Electric Power Company, promises a major leap in the potential to advance materials design. The findings are reported in the December 2024 issue of the journal Nature Computational Science .
Currently, most machine learning models used to describe molecular systems are based on density functional theory (DFT), which provides a quantum mechanical approach to determining the total energy of a molecule or crystal by taking into account the electron density distribution (basically the average number of electrons present in a unit volume around each particular point in the space near the molecule). (Walter Kohn, who co-invented this theory 60 years ago, won the Nobel Prize in Chemistry for it in 1998.) Although the method has been very successful, Lee says it also has some drawbacks. “First, it is not uniformly accurate. Second, it only tells us one thing: the minimum total energy of a molecular system.”
“Couples Therapy” is a lifesaver
His team now relies on another computational chemistry technique rooted in quantum mechanics called cluster boundary theory, or CCSD(T). “This is the gold standard of quantum chemistry,” Li commented. CCSD(T) calculations are much more accurate than DFT calculations and are as reliable as the results we currently get from experiments. The problem is that doing these calculations on a computer is very slow and “it doesn’t scale well. If you double the number of electrons in the system, the computational cost becomes 100 times higher,” he said. That’s why CCSD(T) calculations are usually limited to molecules with a small number of atoms, around 10 or so. Calculations beyond that number take too long.
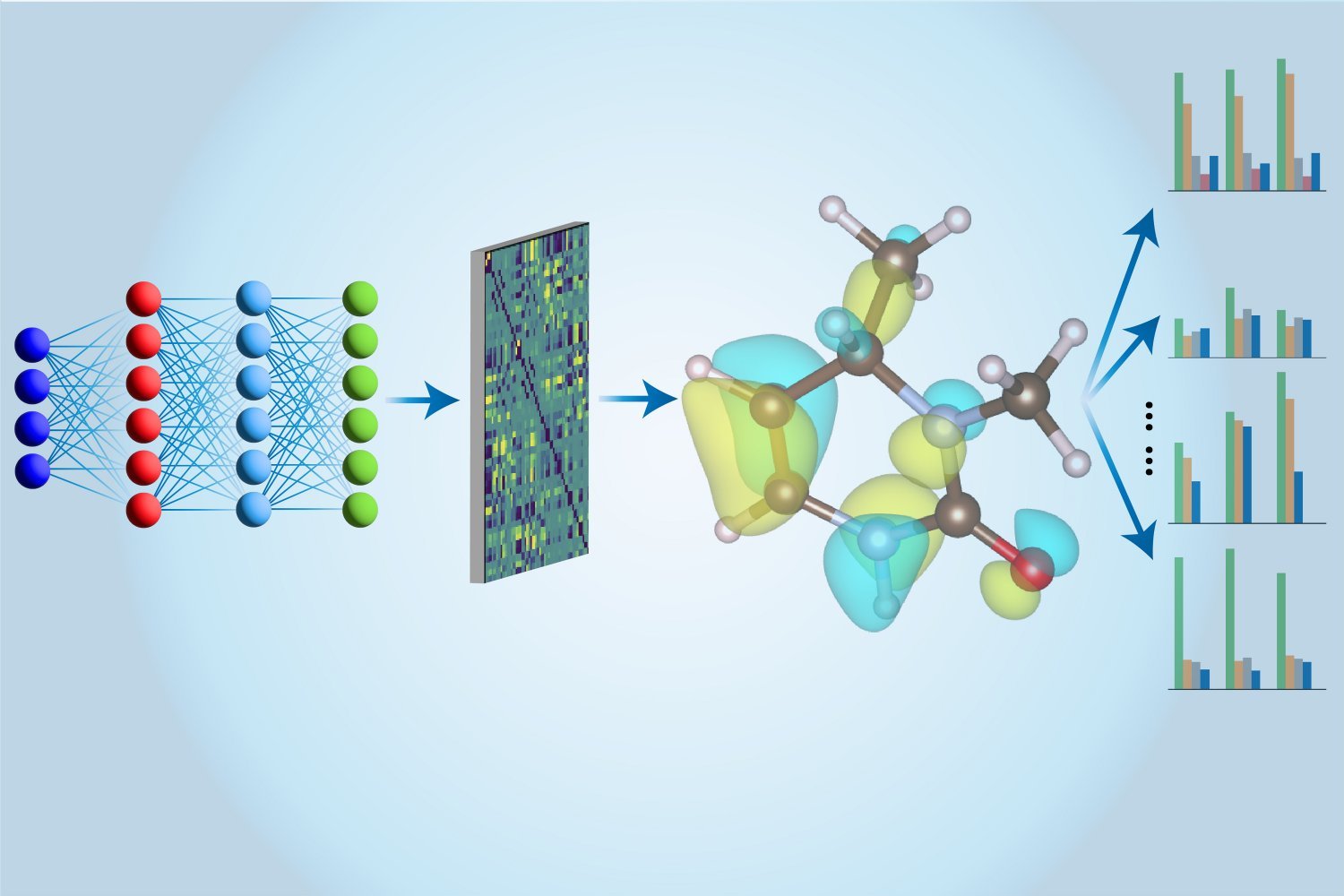
This is where machine learning comes in. The CCSD(T) calculations were first run on a conventional computer, then used to train a neural network using a new architecture that Li and his colleagues specially designed. Once trained, the neural network can take advantage of approximation techniques to perform the same calculations much faster. What’s more, their neural network model can extract more information about the molecules than just their energy. “Previous studies have used different models to evaluate different properties,” says Hao Tang, a doctoral student in materials science and engineering at MIT. “Here, we use just one model to evaluate all these properties, which is why we call it a ‘multitasking’ approach.”
The “multitasking electronic Hamiltonian network,” or MEHnet, reveals various electronic properties, including dipole and quadrupole moments, electronic polarizability, and the optical excitation gap (the amount of energy required to move an electron from the ground state to the lowest excited state). “Excitation distance affects the optical properties of a material,” explains Tan, because it determines the frequencies of light a molecule can absorb. Another advantage of the CCSD-trained model is that it can reveal properties of excited states, not just the ground state. The model can also predict the infrared absorption spectrum of a molecule in relation to its vibrational properties. The vibrations of atoms within a molecule couple with each other to produce various collective behaviors.
The power of their approach relies heavily on the network architecture. Building on the work of MIT Assistant Professor Tess Smit, Tan said the team uses something called an E(3)-equivariant graph neural network, “where nodes represent atoms, and the edges connecting the nodes represent bonds between atoms. We also use custom algorithms that incorporate physical principles related to how quantum mechanics calculates molecular properties directly into our model.”
Test, 1, 2, 3
When tested with known hydrocarbon molecular analyses, the model by Li et al. outperformed the DFT model and closely matched experimental results from published literature.
Qiang Zhu, a materials discovery expert at the University of North Carolina at Charlotte, who was not involved in the research, is impressed with the results so far. “Their method allows for efficient training with small datasets, achieving superior accuracy and computational efficiency compared to existing models,” he said. “This is exciting work that shows the powerful synergy between computational chemistry and deep learning, providing new ideas for developing more accurate and scalable electronic structure methods.”
The MIT-based team first applied the model to small nonmetallic elements that can form organic compounds: hydrogen, carbon, nitrogen, oxygen, and fluorine, then moved on to explore heavier elements silicon, phosphorus, sulfur, chlorine, and even platinum. After training on small molecules, the model can be gradually generalized to larger molecules. “Until now, most calculations have been limited to analyzing hundreds of atoms using DFT and tens of atoms using CCSD(T) calculations,” Li said. “Now we’re talking about dealing with thousands and eventually tens of thousands of atoms.”
Currently, the researchers are still evaluating known molecules, but the model could be used to describe never-before-seen molecules and even predict the properties of hypothetical materials made up of different types of molecules. “The idea is to use our theoretical tools to select promising candidates that meet certain criteria and then propose them to experimentalists for testing,” Tang said.
It all depends on the app
Looking to the future, Zhu is optimistic about potential applications: “This approach has the potential for high-throughput molecular screening,” he said, “a task in which achieving chemical precision is essential to identifying new molecules and materials with desirable properties.”
Demonstrating the ability to analyze large molecules made up of tens of thousands of atoms “will enable us to invent new polymers and materials that can be used in drug design and semiconductor devices,” Li said. Examining heavier transition metal elements could lead to the development of new materials for batteries, an area of urgent need.
In Li’s view, the future is wide open. “It’s no longer just a matter of one region,” he said. “Our ultimate goal is to cover the entire periodic table with CCSD(T)-level accuracy at a lower computational cost than DFT. This will enable us to solve many problems in chemistry, biology, and materials science. At this point, it’s hard to know how wide the scope will be.”
This work was supported by the Honda Research Institute. Hao Tang would like to acknowledge the support of a Mathworks Engineering Fellowship. Portions of the computations in this work were performed on the Matlantis Fast General-Purpose Atomic Simulator, the Texas Center for Advanced Computing, the MIT Supercloud, and the National Energy Research Scientific Computer.